The TACO specification
1. Version and schema
This is version 2.0.0 of the TACO specification, released on November 21, 2025. See Version Compatibility for details on compatibility with previous versions. The specification follows semantic versioning (MAJOR.MINOR.PATCH). Within the same major version, all updates MUST maintain backward compatibility.
2. Overview
The TACO (Transparent Access to Cloud-Optimized datasets) specification defines a comprehensive standard for organizing, storing, and accessing AI-ready Earth Observation (EO) datasets. TACO addresses the fragmentation in current EO dataset practices by providing:
A formal data model that defines the logical structure and relationships of hierarchical geospatial datasets.
A physical storage format optimized for cloud-native access with efficient metadata organization.
A standardized API for creating and reading TACO-compliant datasets across multiple programming languages.
TACO is built on widely-adopted open technologies (GDAL, Apache Parquet) to ensure broad compatibility and long-term maintainability. The specification is designed to support the full lifecycle of AI-ready EO datasets: from creation and distribution to efficient querying and streaming in cloud environments.
3. Design Goals
The TACO specification is guided by the following design principles:
Self-contained and portable: TACO files encapsulate all data and metadata required for interpretation without external dependencies. A TACO dataset can be copied, moved, or archived as a single unit.
Cloud-optimized: The format supports partial reads and efficient byte-range access, enabling streaming workflows without downloading entire datasets. Metadata is organized to minimize HTTP requests in cloud storage scenarios.
Hierarchical by design: TACO natively supports nested dataset structures through the Position-Invariant Tree (PIT) constraint, enabling efficient SQL queries over multi-level hierarchies.
FAIR-compliant: TACO implements the FAIR principles (Findability, Accessibility, Interoperability, Reusability) through standardized metadata, persistent identifiers, clear licensing, and format interoperability.
Language-agnostic: By relying on GDAL Virtual File System (VFS) and Apache Parquet, TACO can be implemented in any language with GDAL bindings (Python, R, Julia, C++, etc.).
Extensible: The specification defines core metadata fields while allowing domain-specific extensions through a formal extension mechanism.
4. Architecture Overview
The TACO specification is structured around three distinct but interconnected layers, each serving a specific purpose in the complete system design (Figure 1):
Data Model Layer (Logical): Defines the abstract structure, relationships, and constraints that govern how datasets are organized. This layer specifies what constitutes a valid TACO dataset independently of how it is physically stored.
Physical Layer (Storage): Defines the concrete serialization format, including byte layout, metadata encoding, and file organization. This layer implements the logical model using established technologies (Apache Parquet, GDAL-compatible formats).
API Layer (Programmatic Access): Provides standardized methods for creating, validating, reading, and querying TACO datasets. This layer abstracts the complexity of the physical format, offering a consistent interface across programming languages.
This separation of concerns enables independent evolution of each layer while maintaining compatibility. For example, the physical layer can adopt new compression algorithms without affecting the data model, and multiple API implementations can coexist (Python, R, Julia, Matlab) as long as they provide the same core methods and functionalities, ensuring that users can switch between languages seamlessly.
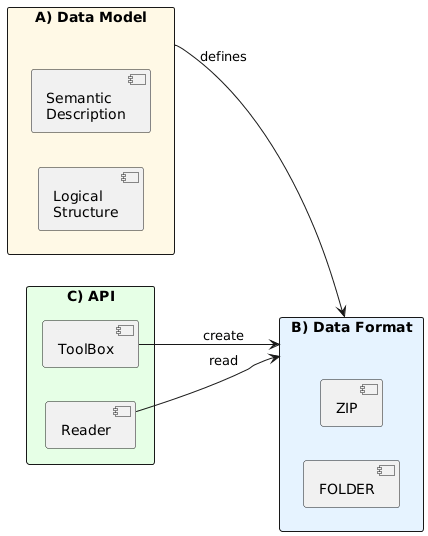 Figure 1: TACO Three-Layer Architecture. The specification separates logical structure (Data Model), concrete serialization (Physical), and programmatic access (API).
Figure 1: TACO Three-Layer Architecture. The specification separates logical structure (Data Model), concrete serialization (Physical), and programmatic access (API).
5. Data Model Layer
5.1. Core Concepts
The TACO data model defines three fundamental abstractions that structure how datasets are organized: SAMPLE, TORTILLA, and TACO. These concepts form a nested hierarchy where each level serves a distinct purpose in representing AI-ready geospatial datasets.
5.2. SAMPLE
A SAMPLE represents the minimal self-contained and smallest indivisible unit. Each SAMPLE encapsulates both data and metadata, providing everything needed to interpret a single instance without external dependencies. Each SAMPLE maintains pointers to both its data and metadata. The metadata itself also contains fields that point to the data, enabling direct access to the underlying files. SAMPLEs are classified into two types based on their content:
FILE: The SAMPLE points to a single data file (e.g., GeoTIFF, JPEG, NetCDF, PDF).
FOLDER: The SAMPLE points to a TORTILLA container, enabling hierarchical structures.
5.3. TORTILLA
A TORTILLA is a collection of SAMPLEs. All SAMPLEs within a TORTILLA MUST have the same metadata schema with consistent types, ensuring that the combined metadata can be represented as a tabular dataframe.
Because a SAMPLE can point to another TORTILLA (FOLDER type), hierarchical dataset structures can be created through recursive nesting. For example, a parent TORTILLA contains SAMPLEs of spatial tiles (FOLDER type), where each spatial tile is itself a TORTILLA containing temporal sequences (FOLDER type), and each temporal sequence is a TORTILLA containing multi-band images (FILE type). This hierarchical organization enables efficient representation of complex geospatial datasets with multiple levels of grouping while maintaining a simple and consistent data model.
5.4. TACO
TACO extends TORTILLA by adding comprehensive dataset-level metadata in a structure called COLLECTION. This metadata provides semantic information about the dataset as a whole, such as identification, provenance, licensing, spatiotemporal coverage, and intended use. A TACO dataset MUST have exactly one root TORTILLA that contains all SAMPLEs.
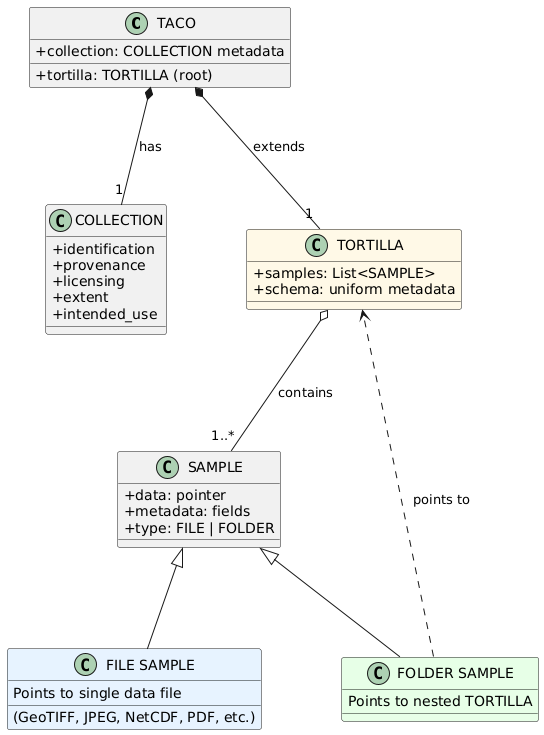
Figure 2: TACO Core Data Model. A SAMPLE is the minimal unit, classified as a FILE (pointing to data files) or a FOLDER (pointing to TORTILLAs). TORTILLA is a collection of SAMPLEs with a uniform metadata schema. TACO extends TORTILLA by adding dataset-level COLLECTION metadata.
5.5. Hierarchical Organization
The PIT is a structural constraint that enables efficient navigation and querying of hierarchical TACO datasets. PIT enforces a regular tree structure where the position of any node uniquely determines its path from the root, eliminating the need for explicit parent-child relationship tracking.
For a dataset to be PIT-compliant, the following rules MUST be enforced:
PIT-1: Structural Isomorphism at Level 0
All SAMPLEs at the root level (level 0) MUST be structurally isomorphic. Two SAMPLEs are considered isomorphic when they satisfy ALL of the following conditions:
- They MUST contain the same number of children
- Children at the same position MUST have identical identifiers (id field)
- Children at the same position MUST have the same type (FILE or FOLDER)
PIT-2: Schema Uniformity Across Levels
All SAMPLEs at the same hierarchy level MUST share an identical metadata schema, including:
- All core metadata fields with matching data types
- All extension fields with matching data types
- Identical field ordering (optional but recommended)
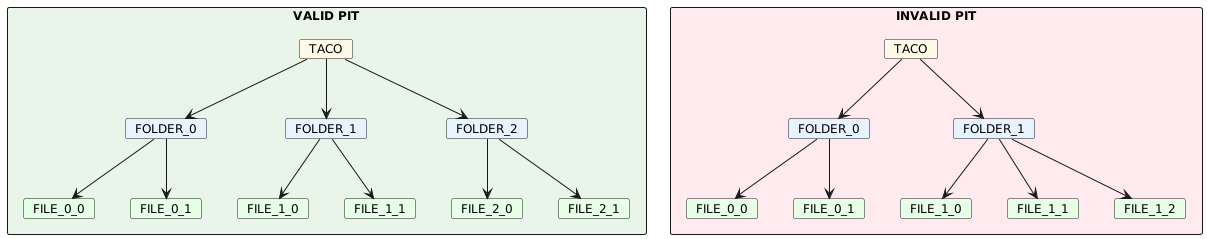 Figure 3: Valid PIT (left) has uniform structure: all FOLDERs contain 2 FILEs. Invalid PIT (right) has different child counts: FOLDER_0 has 2 FILEs, FOLDER_1 has 3 FILEs.
Figure 3: Valid PIT (left) has uniform structure: all FOLDERs contain 2 FILEs. Invalid PIT (right) has different child counts: FOLDER_0 has 2 FILEs, FOLDER_1 has 3 FILEs.
Padding for irregular structures: When datasets have missing observations, placeholder SAMPLEs with identifiers __TACOPAD__N are inserted to maintain PIT compliance.
5.6. Metadata Schema
The TACO data model defines metadata at two levels: SAMPLE-level metadata describing individual units, and TACO-level metadata (COLLECTION) describing the dataset as a whole.
5.5.1. SAMPLE-level Fields
Core fields (REQUIRED):
id(String): Unique identifier for the SAMPLE. MUST NOT contain slashes (/,\) or colons (:). MUST NOT start with double underscore (__), which is reserved for padding.type(String): SAMPLE type. MUST be eitherFILEorFOLDER.path(String | Null): Location of the data. For FILE SAMPLEs, it contains the file path. For FOLDER SAMPLEs, contains a pointer to a TORTILLA.
Extension fields:
SAMPLEs can be extended with domain-specific metadata through the extension mechanism. Extensions add additional fields using namespace prefixes (e.g., stac:* or stats:*). All SAMPLEs within a TORTILLA MUST have identical metadata schemas, including all extension fields.
TORTILLA provides an efficient mechanism for computing extension metadata over sets of SAMPLEs. When metadata computation benefits from accessing the entire dataset (e.g., statistical aggregations, spatial indexing, batch processing), TORTILLA extensions compute fields across all contained SAMPLEs simultaneously rather than individually.
5.5.2. TACO-level Fields (COLLECTION)
Core fields (REQUIRED):
id(String): Unique persistent identifier for the dataset. MUST be lowercase alphanumeric with underscores and hyphens allowed.taco_version(String): TACO specification version (e.g., "2.0.0").dataset_version(String): Version of the dataset itself.description(String): Human-readable description of the dataset.licenses(List[String]): Dataset license(s). SPDX identifiers are RECOMMENDED.providers(List[Object]): Persons or organizations who created the dataset. Each object containsname,organization,email, androlefields.tasks(List[String]): Machine learning tasks the dataset supports (e.g.,["semantic-segmentation"],["classification"]).
Optional fields:
title(String): Short title for the dataset (max 250 characters).curators(List[Object]): Persons responsible for converting the dataset to TACO format. Same structure asproviders.keywords(List[String]): Descriptive keywords for searchability.extent(Object): Spatial and temporal coverage. Containsspatial(bounding box[min_lon, min_lat, max_lon, max_lat]in WGS84) andtemporal(ISO 8601 datetime strings[start, end]). This field can be auto-calculated from SAMPLE-level metadata if the user defines the STAC extension.
Extension fields:
TACO-level extensions add dataset-wide metadata using the same namespace convention as SAMPLE-level extensions.
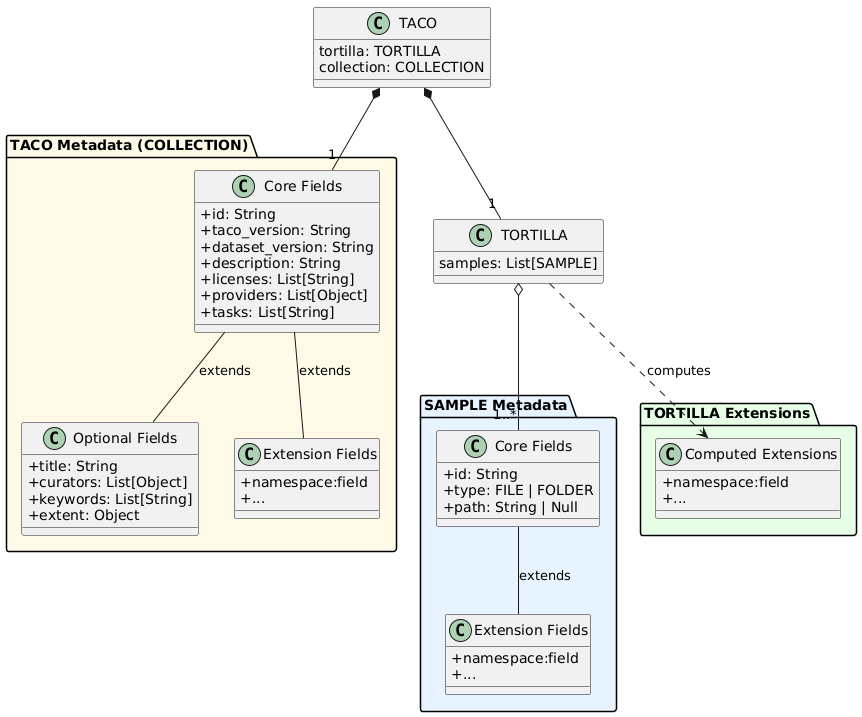 Figure 4: TACO Metadata Schema Overview. SAMPLE metadata consists of core fields (id, type, path) and extension fields using namespace prefixes. TORTILLA provides efficient computation of extensions over collections of SAMPLEs. TACO metadata (COLLECTION) includes core fields for dataset identification, optional fields for additional context, and extension fields for domain-specific metadata. All levels use the same namespace-based extension mechanism.
Figure 4: TACO Metadata Schema Overview. SAMPLE metadata consists of core fields (id, type, path) and extension fields using namespace prefixes. TORTILLA provides efficient computation of extensions over collections of SAMPLEs. TACO metadata (COLLECTION) includes core fields for dataset identification, optional fields for additional context, and extension fields for domain-specific metadata. All levels use the same namespace-based extension mechanism.
6. Physical Layer
6.1. Directory Structure
TACO datasets follow a standardized directory structure consisting of three main components:
DATA/: Contains the hierarchical organization of data files and folders. Each SAMPLE of type FILE is stored as a physical file, while each SAMPLE of type FOLDER is represented as a subdirectory containing its own set of SAMPLEs. This structure mirrors the logical hierarchy defined in the data model.
METADATA/: Contains consolidated metadata for efficient querying across the entire dataset. Metadata is organized into separate files per hierarchy level: level0.parquet, level1.parquet, etc. Each file contains the complete metadata for all SAMPLEs at that specific depth in the hierarchy.
COLLECTION.json: This file contains the dataset-level metadata as a UTF-8 encoded JSON document. It includes all core and optional fields defined in the TACO metadata schema, as well as any TACO-level extensions.
Example 1 - Flat structure (single level):
dataset/
├── DATA/
│ ├── image_001.tif
│ ├── image_002.tif
│ └── image_003.tif
├── METADATA/
│ └── level0.parquet
└── COLLECTION.jsonExample 2 - Three-level hierarchy (scenes -> components -> files):
dataset/
├── DATA/
│ ├── scene_001/
│ │ ├── __meta__
│ │ ├── label.json
│ │ └── imagery/
│ │ ├── __meta__
│ │ ├── before.tif
│ │ └── after.tif
│ ├── scene_002/
│ │ ├── __meta__
│ │ ├── label.json
│ │ └── imagery/
│ │ ├── __meta__
│ │ ├── before.tif
│ │ └── after.tif
├── METADATA/
│ ├── level0.parquet
│ ├── level1.parquet
│ └── level2.parquet
└── COLLECTION.jsonInvalid structure (PIT violation - mixed types at same level):
dataset/
├── DATA/
│ ├── tile_001/ <- FOLDER
│ │ └── __meta__
│ │ └── file_a
│ ├── tile_002/ <- FOLDER
│ │ └── __meta__
│ │ └── file_a
│ └── tile_003 <- FILE (BREAKS PIT!)
├── METADATA/
│ └── level0.parquet
└── COLLECTION.jsonThis violates PIT because level 0 contains both FOLDER SAMPLEs (tile_001, tile_002) and FILE SAMPLE (tile_003). All SAMPLEs at the same level MUST have the same type; this structure is invalid.
6.2. Dual Metadata System
TACO implements a dual metadata strategy to optimize for different access patterns:
Consolidated Metadata (METADATA/levelX.parquet)
Consolidated metadata enables efficient SQL queries across the entire dataset without traversing the directory hierarchy. Each file MUST contain all SAMPLE metadata at its corresponding hierarchy level, including core fields (id, type, path), extension fields, and the automatically generated internal:parent_id field that links each SAMPLE to its parent's index in the previous level.
Consolidated metadata is lazy-loaded at dataset initialization for query planning and execution. This organization enables operations like filtering by spatiotemporal extent, aggregating statistics, or selecting specific SAMPLEs based on arbitrary predicates.
Local Metadata (DATA/folder/meta.parquet)
Local metadata enables fast navigation within a specific folder without loading the entire dataset's metadata. Each __meta__ parquet file MUST contain metadata only for the direct children of that specific folder, representing a complete metadata snapshot for one TORTILLA container. In practical terms, a TORTILLA always maps to a directory that MUST contain a __meta__ file with its children's metadata.
Local metadata is read on demand when navigating into a specific folder or querying a subset of the hierarchy. This organization enables efficient local operations without the overhead of loading global metadata, supporting workflows that process individual folders or subtrees independently.
The dual system allows TACO readers to choose the optimal metadata source based on the query: consolidated metadata for global operations, local metadata for hierarchical navigation.
6.3. ZIP Format
TACO implements a cloud-optimized version of the ZIP format. The standard ZIP format requires scanning the Central Directory located at the end of the file to access internal files. TACO ZIP solves this by including a TACO_HEADER at byte position 0 that contains direct offsets to all metadata files and COLLECTION.json, enabling immediate access without Central Directory traversal.
All files in TACO ZIP archives use STORE mode (compression method = 0), meaning no compression is applied. This design choice enables efficient byte-range access in cloud storage scenarios, GDAL Virtual File System compatibility, and parallel random access patterns without decompression overhead.
When a ZIP container is created, two additional metadata columns MUST be generated: internal:offset indicating the byte offset of each SAMPLE within the ZIP file, and internal:size indicating the length in bytes. These columns enable direct access to individual SAMPLEs without decompressing the entire archive. The ZIP format is immutable; updating metadata requires recreating the entire ZIP file.
6.4. FOLDER Format
The FOLDER format stores TACO datasets as a directory structure. Files remain directly accessible for inspection and modification without requiring extraction or decompression. This format is suitable for development environments and datasets requiring frequent updates.
FOLDER format supports efficient metadata updates through Content-Defined Chunking (CDC). CDC divides parquet files into variable-sized chunks based on content rather than fixed positions, typically using algorithms such as FastCDC/Rabin–Karp with average chunk sizes around 64KB. The storage backend maintains a content-addressed store where each chunk is identified by its cryptographic hash, enabling automatic deduplication of identical chunks across versions.
When metadata is updated, only chunks containing modified data need to be transferred to storage. The storage backend reconstructs files by assembling the appropriate chunks based on their hash references. This approach reduces transfer size proportionally to the magnitude of changes rather than total file size, providing significant efficiency improvements for incremental updates to large Parquet metadata files.
Server support for CDC and chunk-level deduplication is required to achieve these performance characteristics. To our best knowledge, Hugging Face's XET storage backend is currently the only public platform that provides this infrastructure for scientific datasets.
6.5. TACOCAT File
TACOCAT is a consolidated file (__TACOCAT__) that addresses the operational challenges of managing large collections of individual TACO ZIP files. When datasets are distributed as hundreds or thousands of separate ZIP containers, querying across the collection requires opening each file individually to access its metadata. TACOCAT solves this by consolidating all metadata from multiple ZIP files into a single high-performance container.
The format extracts and merges all METADATA/levelX.parquet files from source ZIP containers into consolidated Parquet files, one per hierarchy level. An additional column internal:source_file is added to track the original ZIP filename for each SAMPLE. This enables queries across the entire collection using a single DuckDB connection without filesystem traversal or individual ZIP file access.
TACOCAT uses a fixed 128-byte binary header containing the format magic number, version identifier, maximum hierarchy depth, and a 7-entry index table. The index stores byte offsets and sizes for up to 6 consolidated metadata levels plus the merged COLLECTION.json. Data sections begin at byte 128 and are stored sequentially. The consolidated COLLECTION.json preserves the PIT schema from source datasets and includes provenance metadata listing all source ZIP filenames.
When reading SAMPLEs, the GDAL VSI path is constructed as /vsisubfile/{offset}_{size},{base_path}{source_file} where offset and size reference the original position within the source ZIP file identified by source_file. This allows direct byte-range access to data without extracting or opening intermediate containers.
6.6. TACOLLECTION File
Tacollection is a global metadata consolidation mechanism (TACOLLECTION.json) for datasets distributed across multiple TACO ZIP files. When large datasets are partitioned (e.g., by region or time period), each partition contains independent COLLECTION.json metadata. Tacollection merges these into a unified global view.
The consolidation validates that all partitions share identical taco:pit_schema and taco:field_schema structures. Sample counts are summed to compute the total dataset size. Spatial extents merge into a global bounding box, while temporal extents span from the earliest start to the latest end across all partitions.
The taco:sources field preserves each partition's individual extents, enabling query routing: users can identify which files contain relevant data for a specific region or time window without opening every partition. The field stores partition count, IDs, filenames, and individual spatial/temporal coverage:
{
"taco:sources": {
"count": 3,
"ids": ["europe_2023", "asia_2023", "americas_2023"],
"files": ["europe.tacozip", "asia.tacozip", "americas.tacozip"],
"extents": [
{
"file": "europe.tacozip",
"id": "europe_2023",
"spatial": [-10, 30, 45, 70],
"temporal": ["2023-01-01T00:00:00Z", "2023-12-31T23:59:59Z"]
},
{
"file": "asia.tacozip",
"id": "asia_2023",
"spatial": [60, -10, 150, 55],
"temporal": ["2023-01-01T00:00:00Z", "2023-12-31T23:59:59Z"]
},
{
"file": "americas.tacozip",
"id": "americas_2023",
"spatial": [-170, -55, -35, 75],
"temporal": ["2023-01-01T00:00:00Z", "2023-12-31T23:59:59Z"]
}
]
}
}7. API Layer
The TACO API provides two tools: TacoToolbox for creating datasets and TacoReader for querying them.
TacoToolbox (Writer) is Python-only for ease of maintenance, not technical necessity.
TacoReader (Reader) is designed to be easily implemented in any language with a DataFrame interface, object-oriented classes, a lazy Parquet reader with query pushdown support, and GDAL bindings for data access. This makes TacoReader viable in R, Rust, Julia, C++, and other scientific computing languages.
All TacoReader and TacoToolbox MUST conform to the core API specification defined below. Additional features beyond the specification MUST be implemented in separate modules to prevent fragmentation.
7.1. TacoToolbox (Writer API)
TacoToolbox provides functionality for creating, converting, and validating TACO datasets. The API is organized around three core concepts: SAMPLE (minimal unit), TORTILLA (set of SAMPLEs), and TACO (complete dataset).
7.1.1. Data Model Classes
datamodel.Sample
Represents the minimal self-contained unit in a TACO dataset. Each SAMPLE encapsulates data and metadata.
Sample(
id: str, # Unique identifier
type: "FILE" | "FOLDER" | "auto", # Type inference by default
path: Path | bytes | TORTILLA, # Data location
temp_dir: Path = None, # For bytes conversion
**metadata # Extension fields
)Type Inference: When type="auto" (default), type is automatically inferred from path. Path or bytes become a FILE, TORTILLA becomes a FOLDER. After validation, the type is always FILE or FOLDER.
Bytes Support: When passing bytes as a path, temporary files are created automatically. Use the temp_dir parameter to specify the location for large datasets. Files are cleaned up on garbage collection or an explicit cleanup() call.
datamodel.Tortilla
Collection of SAMPLEs with uniform metadata schema. Validates PIT constraints.
Tortilla(
samples: list[Sample],
pad_to: int = None, # Auto-pad to divisible length
strict_schema: bool = True # Enforce schema uniformity
)Auto-Padding: The pad_to parameter automatically adds padding to SAMPLEs to make the total length divisible by the specified value. Padding SAMPLEs use ID prefix __TACOPAD__ with empty bytes, creating zero-byte temporary files.
Schema Modes: With strict_schema=True, all SAMPLEs MUST have identical metadata schemas. With strict_schema=False, heterogeneous schemas are allowed with automatic None-filling for missing columns.
datamodel.Taco
Complete dataset extending TORTILLA with collection-level metadata.
Taco(
tortilla: Tortilla,
id: str,
dataset_version: str,
description: str,
licenses: list[str],
providers: list[Contact],
tasks: list[TaskType],
taco_version: str = "2.0.0",
title: str = None, # Max 250 characters
curators: list[Contact] = None,
keywords: list[str] = None,
extent: Extent = None # Auto-calculated from STAC/ISTAC
)Extent Auto-Calculation: If extent is not provided, TACO searches incrementally through hierarchy levels for STAC or ISTAC metadata and computes spatial and temporal extents automatically. Defaults to global extent if no spatiotemporal metadata found.
7.1.2. Extension System
The extension system enables adding computed or declarative metadata to SAMPLEs, TORTILLAs, and TACOs through a unified extend_with() interface.
SAMPLE Extensions
SAMPLE extensions add metadata fields computed from the SAMPLE's data. Extensions can be computational (implementing SampleExtension abstract class) or declarative (dict, DataFrame, Pydantic model).
class SampleExtension(ABC):
schema_only: bool = False # Return None values preserving schema
@abstractmethod
def get_schema(self) -> dict[str, pl.DataType]:
"""Return expected DataFrame schema."""
@abstractmethod
def _compute(self, sample: Sample) -> pl.DataFrame:
"""Compute metadata, return single-row DataFrame."""The schema_only parameter enables schema-first workflows where structure is defined without computation cost. When True, _compute() is never called and None values are returned with the proper schema.
Extension fields are added directly to the SAMPLE model and tracked in _extension_schemas for proper DataFrame serialization. Key validation ensures extension fields use valid formats: alphanumeric plus underscore, optionally with a colon for namespaces.
TORTILLA Extension
TORTILLA extensions add columns to the metadata DataFrame computed across all SAMPLEs in the collection.
class TortillaExtension(ABC):
schema_only: bool = False
@abstractmethod
def get_schema(self) -> dict[str, pl.DataType]:
"""Return expected schema."""
@abstractmethod
def _compute(self, tortilla: Tortilla) -> pl.DataFrame:
"""Compute metadata, return DataFrame with one row per sample."""The returned DataFrame MUST have exactly one row per SAMPLE. Column conflicts with existing metadata are rejected. Extensions are applied via horizontal concatenation.
TACO Extensions
TACO extensions add dataset-level computed metadata.
class TacoExtension(ABC):
@abstractmethod
def get_schema(self) -> dict[str, pl.DataType]:
"""Return schema."""
@abstractmethod
def _compute(self, taco: Taco) -> pl.DataFrame:
"""Compute metadata, return single-row DataFrame."""Extension data is added as instance attributes directly to the TACO object.
7.1.3. Core Functions
create(taco, output, output_format="auto", **kwargs)
Primary function for writing TACO datasets to disk. Supports both ZIP and FOLDER container formats. Format auto-detected from extension (.zip/.tacozip -> ZIP, else -> FOLDER). Parquet parameters passed through kwargs (compression, compression_level, row_group_size).
create_tacocat(inputs, output, **parquet_kwargs)
Consolidates multiple ZIP containers into a single high-performance file. Merges all metadata into consolidated Parquet files with internal:source_file column tracking origin. Validates schema consistency across datasets. Generates a fixed-name __TACOCAT__ file with a 128-byte binary header containing format magic, version, maximum depth, and a 7-entry index table.
create_tacollection(inputs, output, validate_schema=True)
Creates global TACOLLECTION.json from multiple datasets. Validates PIT schema structure and field schema consistency across all inputs. Sums sample counts in taco:pit_schema hierarchy, computing total dataset size. Merges spatial extents into a global bounding box covering all partitions, and merges temporal extents into a global time range spanning the earliest start to the latest end. Preserves individual partition extents in taco:sources field for query routing and visualization. Uses the first dataset as a base for other metadata fields. Always generates a standard-named TACOLLECTION.json file in the output directory.
export(dataset, output, output_format="auto", quiet=False, limit=100, temp_dir=None, **kwargs)
Exports filtered subsets of existing TACO datasets. Format auto-detected from extension (.zip/.tacozip -> ZIP, else -> FOLDER). Uses async I/O with configurable concurrency limit for remote data sources. Preserves hierarchical structure while updating collection metadata with subset provenance tracking via taco:subset_of and taco:subset_date fields. Parquet parameters passed through kwargs.
7.1.4. Format Conversion
folder2zip(input, output, quiet=False, temp_dir=None, **kwargs)
Converts FOLDER format to ZIP format. Reads existing metadata from FOLDER structure, including COLLECTION.json and consolidated metadata. Reconstructs local metadata from consolidated metadata for __meta__ file generation. Scans the DATA directory for physical files. Uses ZipWriter to create a ZIP container with bottom-up offset calculation, regenerating all __meta__ files with internal:offset and internal:size columns. Parquet parameters passed through kwargs.
zip2folder(input, output, limit=100, quiet=False)
Converts ZIP format to FOLDER format. Async operation with configurable limit for parallel file extraction. Uses ExportWriter wrapper providing progress bars unless quiet mode is enabled.
7.1.5. Logging Control
verbose(level=True|"debug"|False)
Controls logging verbosity across all TacoToolbox operations. Level True or "info" shows standard progress, including file counts and operations. Level "debug" shows detailed internal operations, including offset calculations and metadata processing. Level False disables all logging output. Settings apply globally to all subsequent operations until changed.
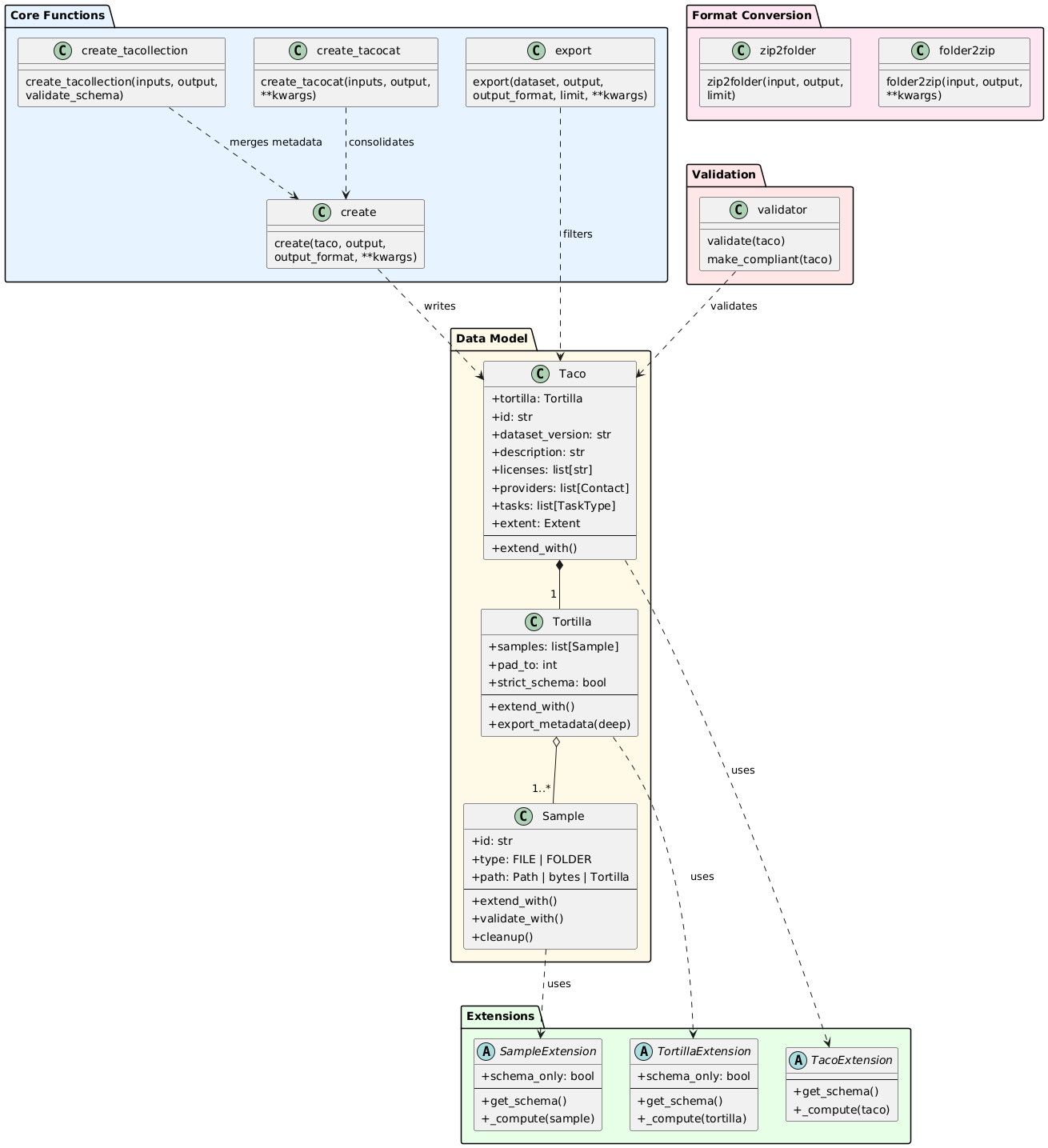 Overview of TacoToolbox writer API
Overview of TacoToolbox writer API
7.2. TacoReader (Reader API)
TacoReader provides functionality for loading and querying TACO datasets through a lazy query interface with SQL support. The API is organized around three core concepts: TacoDataset (lazy query interface), TacoDataFrame (materialized results with navigation), and PITSchema (structural validation).
7.2.1. Loading Datasets
load(path, base_path=None)
Main entry point for loading TACO datasets with automatic format detection. Supports single paths or lists with automatic concatenation if schemas are compatible.
load(
path: str | list[str], # Single path or list for auto-concat
base_path: str = None # TACOCAT only: override ZIP locations
)Format Detection: Automatically detects format from path structure. TACOCAT is identified by a fixed filename __TACOCAT__. ZIP identified by .tacozip or .zip extensions. FOLDER identified by directory structure (default when no extension match).
List Handling: When provided a list of paths, loads each dataset and automatically concatenates if the schemas are compatible. Empty lists raise ValueError. Single-element lists are unwrapped to a simple load operation.
TACOCAT Base Path Override: The base_path parameter enables loading TACOCAT index files separate from source ZIP files. When specified, reconstructs all query views using the new base path for /vsisubfile/ VSI path construction. Useful for distributed storage where the metadata index and data files reside in different locations.
7.2.2. TacoDataset - Lazy Query Interface
TacoDataset provides a STAC-like metadata container with a lazy query interface supporting SQL operations. All operations create views without materializing data until the data attribute is accessed.
Public Metadata (STAC-like):
id,version,description: Dataset identificationtasks,extent,providers,licenses: Dataset characteristicstitle,curators,keywords: Optional descriptive metadatapit_schema: PIT structural schemafield_schema: Column schemas by hierarchy levelcollection: Complete COLLECTION.json content
Attributes
The data attribute materializes the current view into a TacoDataFrame by executing the query and loading results into memory. This is where lazy evaluation ends and data becomes available for operations.
The field_schema attribute returns a column schema dictionary from taco:field_schema in the collection metadata, listing available columns and their types at each hierarchy level.
The collection attribute returns complete COLLECTION.json content with all metadata as a dictionary.
SQL Interface
The sql(query) method executes an SQL query, creating a new TacoDataset instance with a lazy view. Query not executed immediately, only view created. Must use data as the table name, which implementations auto-replace with the current view name, enabling query chaining. Query chaining example in Python:
ds1 = dataset.sql("SELECT * FROM data WHERE cloud_cover < 10")
ds2 = ds1.sql("SELECT * FROM data WHERE country = 'Peru'")
result = ds2.data # Executes combined querySTAC-Style Filtering
filter_bbox(minx, miny, maxx, maxy, geometry_col="auto", level=0)filters by spatial bounding box using spatial query capabilities. Whengeometry_col="auto", searches columns in priority order:istac:geometry,stac:centroid,istac:centroid. Whenlevel > 0, filters level0 samples based on children's geometry using cascading JOINs.filter_datetime(datetime_range, time_col="auto", level=0)filters by temporal range using native Parquet TIMESTAMP columns. Accepts string range"2023-01-01/2023-12-31", single datetime object, or tuple of datetime objects. Whentime_col="auto", searches columns in priority order:istac:time_start,stac:time_start. Always usestime_start. Whenlevel > 0, filters level0 samples based on children's timestamps.
Cascading Joins: Multi-level filtering uses INNER JOINs: level0 -> level1 -> level2 -> target_level using internal:parent_id foreign keys. Returns DISTINCT level0 samples whose descendants match filter criteria. JOIN strategy varies by format: ZIP/FOLDER uses parent_id referencing parent's ID string, TACOCAT uses parent_id as local index plus internal:source_file for disambiguation across multiple source ZIPs.
7.2.3. TacoDataFrame - Hierarchical Navigation
TacoDataFrame wraps DataFrame with TACO-specific functionality for hierarchical navigation. Provides read() method for traversing nested structures.
Protected Columns: Critical columns that MUST NOT be removed or altered as they are essential for hierarchical navigation. The table below lists all protected columns with their purposes and format support. All internal:* columns are created by TacoToolbox during dataset generation, except internal:gdal_vsi, which is dynamically constructed by TacoReader backends.
| Column | Purpose | Format Support |
|---|---|---|
id | Unique sample identifier within parent scope. Used as a primary key for lookups, references, and read() navigation by name. Must be unique among siblings. | ZIP, FOLDER, TACOCAT |
type | Sample type discriminator. Values: FILE (leaf node with data) or FOLDER (intermediate node with children). Determines navigation behavior and metadata structure. | ZIP, FOLDER, TACOCAT |
internal:parent_id | Foreign key referencing the parent sample's position in the previous level. Enables relational queries across hierarchy (level0 → level1 → level2). Value is an integer index in the parent level DataFrame. | ZIP, FOLDER, TACOCAT |
internal:offset | Byte offset in container file where sample data begins. Used to construct /vsisubfile/ VSI paths for GDAL byte-range access without extraction. | ZIP, TACOCAT |
internal:size | Size in bytes of sample data in container. Combined with offset for /vsisubfile/{offset}_{size},{path} VSI path construction. | ZIP, TACOCAT |
internal:gdal_vsi | Complete GDAL Virtual File System path for direct raster access. Format varies by container: /vsisubfile/... for ZIP/TACOCAT, filesystem paths for FOLDER. Dynamically constructed by reader backends. | ZIP, FOLDER, TACOCAT |
internal:source_file | Original source ZIP filename in TACOCAT consolidated datasets. Disambiguates samples from multiple source files sharing the same parent_id values. | TACOCAT only |
internal:relative_path | Relative path from DATA/ directory. Used for filesystem navigation in FOLDER format and path reconstruction in all formats. Format: {parent_path}/{id} or direct {id} for level0. | ZIP, FOLDER, TACOCAT |
TacoDataFrame wraps DataFrame with TACO-specific functionality for hierarchical navigation. Provides read() method for traversing nested structures.
Hierarchical Navigation
The read(key) method navigates to the child level by position (int) or ID (str). FILE samples return GDAL VSI path as a string, enabling direct raster access. FOLDER samples read __meta__ file and return TacoDataFrame with children.
Path construction varies by format:
ZIP:
/vsisubfile/{offset}_{size},{zip_path}enables GDAL byte-range access without extractionFOLDER: Direct filesystem paths
{parent_path}/{relative_path}or{parent_path}/{id}TACOCAT:
/vsisubfile/{offset}_{size},{base_path}{source_file}enabling byte-range access across multiple source ZIPs
Navigation example:
tdf = dataset.data
child = tdf.read(0) # By position
child = tdf.read("sample_01") # By ID
vsi_path = tdf.read(5) # Returns str if FILE7.2.4. Concatenation
concat(datasets, column_mode="intersection")
Concatenates multiple TacoDataset instances into a single dataset with a lazy query interface. Creates consolidated metadata views without disk materialization.
Column Modes:
intersectionmode (default, safest) keeps only columns present in ALL datasets. Drops columns existing only in some datasets with a warning showing which columns were dropped and from which datasets. Suitable when datasets have slight schema variations and only common columns are needed.fill_missingmode keeps all columns from all datasets, filling missing columns with NULL. Warns showing which columns are being filled and in which datasets. Suitable when all columns are needed, even if some datasets lack them.strictmode fails if columns differ between datasets. Raises ValueError with a detailed breakdown showing exact schema per dataset, columns only in some datasets, and columns common to all. Suitable for production pipelines requiring schema homogeneity.
View Construction: Creates query views combining all datasets with internal:source_file column tracking origin dataset. Filters padding samples (__TACOPAD__N) from user-facing views. Constructs internal:gdal_vsi paths per format for data access.
7.2.5. Backend System
Three backends handle format-specific loading strategies, sharing a common interface through an abstract base class.
ZipBackend reads
.tacozipformat using TACO_HEADER at file start for direct offset-based access without full ZIP extraction. Constructs VSI paths as/vsisubfile/{offset}_{size},{zip_path}, enabling GDAL byte-range access.FolderBackend reads the directory structure with loose files. Level 0 constructs paths as
{root}DATA/{id}, level 1+ uses{root}DATA/{internal:relative_path}.TacoCatBackend reads consolidated format from multiple source ZIPs. Parses a fixed 128-byte binary header containing format magic, version, max depth, and a 7-entry index table. Loads entire file to memory (typically <1GB metadata-only). Constructs VSI paths as
/vsisubfile/{offset}_{size},{base_path}{source_file}wheresource_filecolumn identifies origin ZIP.
7.2.6. Logging Control
verbose(level=True|"debug"|False)
Controls logging verbosity across all TacoReader operations. Level True or "info" shows standard progress, including file loading, backend operations, and view creation. Level "debug" shows detailed internal operations including DuckDB queries, offset calculations, and memory operations. Level False disables all logging output. Settings apply globally to all subsequent operations until changed.
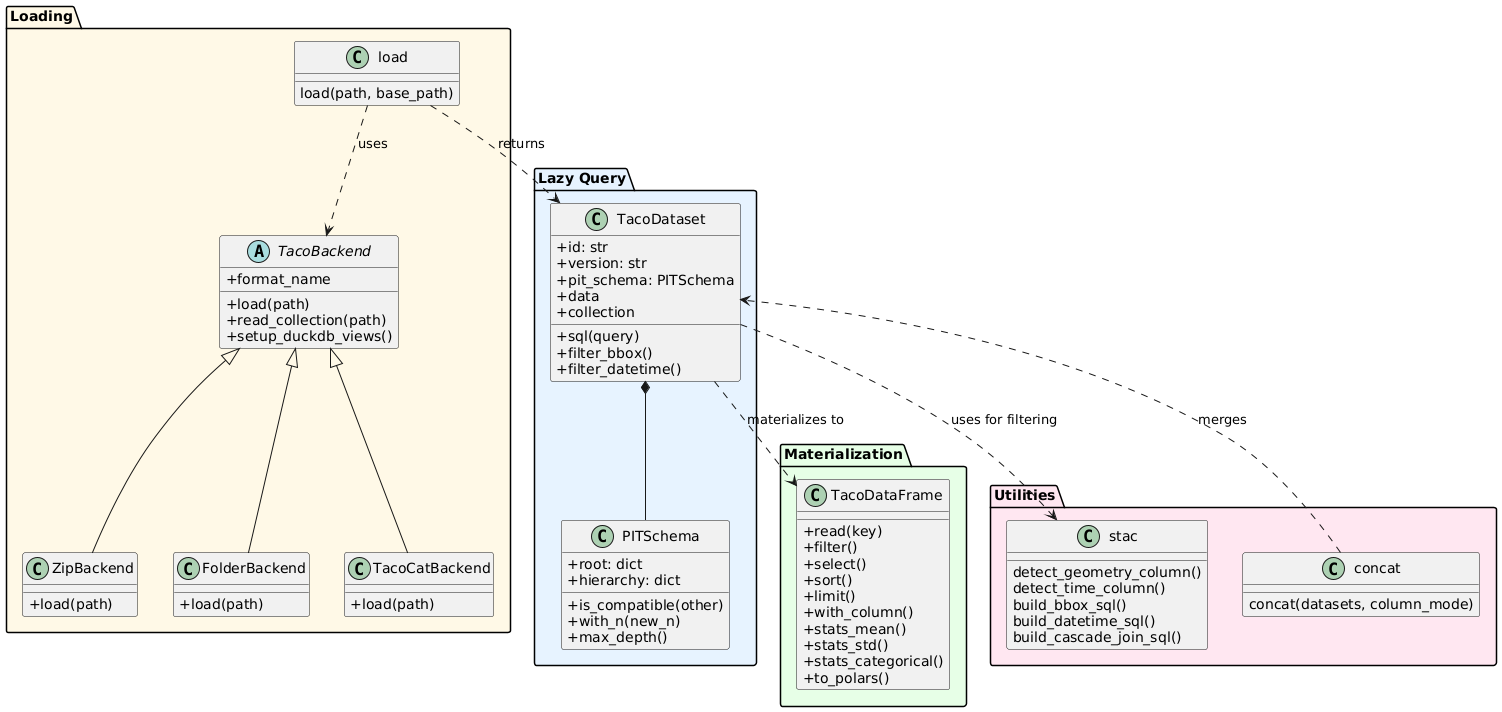 Overview of TacoReader writer API
Overview of TacoReader writer API
8. Version Compatibility
TACO v2.0.0 is NOT backward compatible with v1.x datasets. Existing v1.x datasets MUST be migrated or recreated to work with v2.0.0 tools. Key breaking changes include:
PIT Enforcement: v2.0.0 requires strict PIT compliance at all hierarchy levels. v1.x allowed heterogeneous structures where siblings could differ in child count or organization.
Metadata Schema: Core metadata fields have been renamed, removed, or changed types. Field names now use namespace prefixes (e.g.,
stac:*,istac:*) for clarity and extensibility.Extension Mechanism: v2.0.0 introduces a unified extension system with
extend_with()interface applicable to SAMPLEs, TORTILLAs, and TACOs. v1.x used separate, incompatible extension approaches for each level.Container Formats: v2.0.0 uses standard ZIP archives and directory structures (FOLDER) accessible with generic tools. v1.x relied on a custom BLOB format requiring specialized readers.
Dual Metadata System: v2.0.0 implements both consolidated metadata (
METADATA/levelX.parquet) for SQL queries and local metadata (__meta__files) for navigation. v1.x supported only local metadata, requiring full traversal for dataset-wide operations.API Redesign: TacoToolbox and TacoReader have been completely redesigned with breaking changes to function signatures and class interfaces.
Annex A: Glossary
Container Format: Physical storage structure for TACO datasets. Two formats supported: ZIP (compressed archive) and FOLDER (directory structure).
Consolidated Metadata: Parquet files containing all sample metadata for a hierarchy level. Stored as
METADATA/level0.parquet,METADATA/level1.parquet, etc. Enables efficient SQL queries without traversing individual samples.Extension: Modular mechanism for adding domain-specific metadata to SAMPLEs, or TACOs. Extensions define schema and computation logic, enabling standardized metadata enrichment.
Hierarchical Navigation: Traversal through nested FOLDER samples using
read()method. Each level represents a containment relationship (e.g., country -> region -> scene).Local Metadata: Sample-specific metadata stored in
__meta__files within FOLDER samples. Contains children's metadata for direct filesystem access without consolidated files.PIT (Position-Invariant Tree): Structural constraint ensuring all root samples at the same hierarchy level share identical structure.
Sample: Atomic unit in TACO datasets. Can be a FILE (leaf node with data) or a FOLDER (intermediate node with children). Each sample has a unique ID within the parent scope.
STAC (SpatioTemporal Asset Catalog): Open standard for geospatial metadata. TACO provides STAC-compatible metadata structure while adding hierarchical organization and validation.
TACO: Complete dataset with collection-level metadata (licenses, providers, extent). Contains one TORTILLA with all samples plus descriptive metadata.
TACOCAT: Consolidated format merging multiple TACO ZIP datasets into a single file with a 128-byte binary header and index table. Optimized for querying large collections without opening individual ZIPs.
TORTILLA: Collection of samples with uniform metadata schema. Building block for TACO datasets.
VSI Path: GDAL Virtual File System path format. Enables direct raster access from compressed archives using
/vsisubfile/{offset}_{size},{path}syntax for byte-range reads.
Annex B: History
TACO was born in Valencia, Spain, at the Image and Signal Processing (ISP) group of the Universitat de València. Every Friday, we met at TKOTACO - a one-euro taco restaurant - to talk about series, anime, memes, data, and sometimes new deep learning papers. The regular crew was Julio Contreras, Oscar Pellicer, Simon Donike, Chen Ma (HIT), and me (@csaybar). When David Montero (University of Leipzig) visited, he always joined us.
During one of those Friday meetings, frustrated after spending over a month harmonizing deep learning datasets for cloud detection, we sketched a general solution. We wanted something simple for our problems and ISP datasets. That day, TACOv1 was born - a format with a new BLOB structure and Parquet metadata. The idea was simple: build the infrastructure around GDAL and the DataFrame concept. We converted some datasets to TACOv1, shared the idea with friends, and got valuable feedback.
The format had many flaws, but the core idea was sound. At ESA Living Planet 2025, I had the opportunity to meet several people who shaped TACOv2. Jérémy Anger (Kayrros/Université Paris-Saclay) suggested we abandon the custom BLOB format and use ZIP instead - this became a turning point. Now that we have tacozip, we're happy we listened to his advice. Around the same event, conversations with Mikolaj Czerkawski (Asterisk) about MajorTOM helped us address another critical challenge: designing strategies for large datasets where metadata becomes a bottleneck. His insights on data access patterns influenced much of our lazy loading architecture. Nils Lehmann and Adam Stewart (University of Munich, TorchGeo) brought years of experience working with deep learning datasets, which proved critical for many API design decisions we made.
Recently, Nate Mankovich (ISP) helped us refine the idea from a more mathematical and formal perspective. With his assistance, we defined PIT (Position-Invariant Tree), which became the foundation of TACOv2. Luis Gómez-Chova and Gustau Camps-Valls (ISP) have been discussing what TACO is and should be since day one - many of their ideas are embedded in the code. I'm grateful for their support from the very beginning. They are technically our supervisors, but to us, they feel more like fellow PhD students.
This is the story of TACO - a format born in a one-euro taco restaurant in Valencia. What makes me happiest about this framework is that it emerged around universities, outside all the deep learning hype, among friends laughing about life while coding or writing papers. TACO's code is open source and always will be. Thank you for reading this far.