
The Tortilla specification
The file format behind TACO.
The terms “MUST”, “MUST NOT”, “REQUIRED”, “SHALL”, “SHALL NOT”, “SHOULD”, “SHOULD NOT”, “RECOMMENDED”, “MAY”, and “OPTIONAL” in this document follow the definitions from RFC 2119.
The term “data user” refers to any individual or system accessing the data, “data provider” refers to the entity responsible for offering the data.
Overview
A Tortilla is an open-source, cloud-optimized format for storing large collections of files (referred to as “samples”) in a Binary Large Object (BLOB). It is similar to safetensor and puffin, but designed for seamless compatibility with the Geospatial Data Abstraction Library (GDAL) Virtual File System.
Goals
-
Sample isolation 🛡️ — Each sample is isolated, ensuring that concurrent reads do not interfere with each other. This guarantees consistency and prevents read errors.
-
Speed ⚡ — Tortilla optimizes file scans for both remote and local access by centralizing metadata in the FOOTER, allowing for partial lookups without scanning the entire file.
-
Easy to understand 🧠 — The format is designed with simplicity in mind, making it accessible for developers of all skill levels. Tortilla’s architecture prioritizes clear, concise documentation with a lot of examples and intuitive design for smooth integration and usage.
-
Minimal dependencies 📦 — Tortilla only needs
Apache ArrowandGDALlibraries, making it easy to set up. These libraries are well-supported in many programming languages, such as C++, C#, Go, Java, JavaScript, Julia, MATLAB, Python, R, Ruby, and Rust. -
Nested properties 🌳 — Tortilla supports nested files, allowing data providers to store Tortilla files within other Tortilla files. This feature is especially useful for organizing large, multidimensional datasets.
Format
This is version 0.5.0 of the Tortilla specification. Future versions MUST remain backward compatible with this one. The internal structure of a tortilla file looks like this:
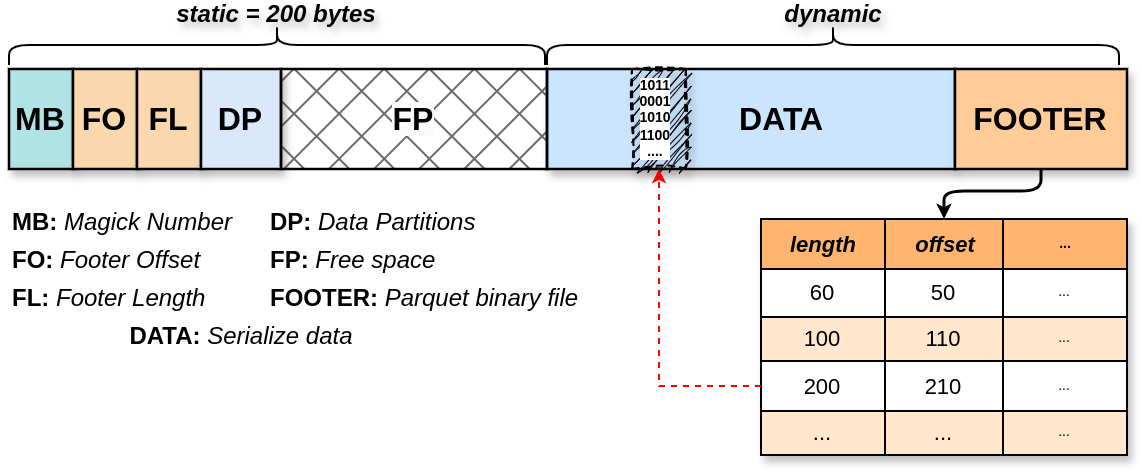
Figure 01: This diagram visually represents the internal structure of a tortilla file, highlighting how data and metadata are organized within the file format.
The Magic Number (MB)
A 2-byte identifier at the start of each file (#y in hex) ensures that the file is recognized as a valid Tortilla format.
int.from_bytes(b'#y', byteorder='little') # 03-10-11 (don't forget)
The Footer Offset (FO)
An 8-byte fixed-size section that indicates where the footer starts within the file. The bytes are stored in little-endian.
The Footer Length (FL)
An 8-byte fixed-size section that specifies the length of the footer in bytes.
The Data Partition (DP)
An 8-byte fixed-size field specifies the number of chunks in a Tortilla file. If it is 1, the file extension MUST be .tortilla. If it is greater than 1, the extension MUST be .part.tortilla, and the chunks MUST be numbered in ascending order. For example, a 100 GB soyuna.tortilla file split into 5 chunks of 20 GB each would result in:
soyuna.0001.part.tortillasoyuna.0002.part.tortillasoyuna.0003.part.tortillasoyuna.0004.part.tortillasoyuna.0005.part.tortilla
Every partition MUST be independently readable. This means that a data user can access a single partition without needing to download all the other partitions.
The Free Space (FP)
A 174-byte fixed-size field specifies the free space remaining at the end of the file’s static section, reserved for future extensions.
The Footer (FOOTER)
The FOOTER is a binary serialized Parquet file that stores TABULAR metadata for each item. Four columns are mandatory:
| Field | Type | Required | Details |
|---|---|---|---|
| tortilla:id | String | Yes | Unique identifier for each item. |
| tortilla:file_format | String | Yes | The format name MUST follow the GDAL naming convention. For example: - GeoTIFF files use the format name GTiff. - JPEG files use the format name JPEG. To get GDAL format names, you can run in the terminal: gdalinfo --formats Additional Supported Formats: - BYTES: Used for data formats not supported by GDAL. - TORTILLA: Used when the file represents a nested Tortilla structure. |
| tortilla:offset | Long | Yes | Byte offset where the item’s data begins in the file. This field is automatically generated by the taco-toolbox. |
| tortilla:length | Long | Yes | Number of bytes that the item’s data occupies. This field is automatically generated by the taco-toolbox. |
| tortilla:data_split | String | No | The data split type. MUST be one of the following: - train: Training data. - test: Testing data. - validation: Validation data. |
To classify a Tortilla file as a stac-tortilla, it must include the following fields:
| Field | Type | Required | Description |
|---|---|---|---|
stac:crs | String | Yes | The Coordinate Reference System (CRS), specified using a recognized authority (EPSG, ESRI or SR-ORG). |
stac:geotransform | Array of Floats | Yes | A 6-element array defining the affine transformation from pixel coordinates to spatial coordinates. The format follows GDAL conventions: [a, b, c, d, e, f], where: - a: Top-left x-coordinate of the upper-left pixel. - b: Pixel width (x-resolution). - c: Row rotation (typically 0 for north-up images). - d: Top-left y-coordinate of the upper-left pixel. - e: Column rotation (typically 0 for north-up images). - f: Negative pixel height (y-resolution, negative for north-up images). This field is essential for georeferencing raster data. |
| stac:raster_shape | 2D array of integers | Yes | The spatial dimensions of the sample, represented as a 2D array shape. |
stac:time_start | Integer | Yes | Timestamp in seconds since UNIX epoch, representing the nominal start time of acquisition. |
stac:time_end | Integer | Yes | Timestamp marking the nominal end of the acquisition or composite period. |
stac:centroid | string | No | This field represents the centroid of the Sample in Well-Known Text (WKT) POINT representation, using the EPSG:4326 coordinate reference system. It is automatically generated by the taco-toolbox. |
Current approaches to building datasets focus primarily on technical and scientific metrics, often overlooking the human impact. To encourage Data providers to consider responsible AI practices, we have adapted the Croissant RAI specification to include optional fields addressing the economic, social, and environmental impacts of the regions surrounding Samples. To classify a Tortilla file as a rai-tortilla, it must include the following fields:
| Field | Type | Required | Details |
|---|---|---|---|
| rai:elevation | Long | yes | Elevation provides critical context about the geographical and environmental conditions of the data collection region. It is RECOMMENDED to measure the rai:elevation as the average in the Sample footprint using the Copernicus Digital Elevation Model (10.5069/G9028PQB). It MUST be in meters. |
| rai:cisi | Float | yes | The Critical Infrastructure Spatial Index (CISI) is a global, grid-based measure of CI proximity. It covers health facilities, educational institutions, water sources, telecommunications, transportation, energy, and waste management. CISI values range from 0 (no CI) to 1 (highest CI intensity). It is RECOMMENDED to calculate rai:cisi as the average within the Sample footprint. For details, see 10.1038/s41597-022-01218-4. |
| rai:gdp | Float | yes | The Gross Domestic Product (GDP) is a critical economic indicator that reflects the economic activity within a region. It is measured in USD per year. It is RECOMMENDED to measure the rai:gdp as the average within the Sample footprint. For details, see 10.1038/sdata.2018.4. |
| rai:hdi | Float | yes | The Human Development Index (HDI) is a composite measure of human well-being. It is calculated based on life expectancy, education, and per capita income indicators. HDI values range from 0 (lowest) to 1 (highest). It is RECOMMENDED to measure the rai:hdi as the average within the Sample footprint. For details, see 10.1038/sdata.2018.4. |
| rai:gmi | Float | yes | The Global human modification index (GMI) is a global, grid-based measure of human modification of terrestrial lands. It is RECOMMENDED to calculate rai:gmi as the average within the Sample footprint. For details, see 10.5194/essd-12-1953-2020. |
| rai:pop | Float | yes | The LandScan Global Population Dataset provides an estimate of the population at 1 km resolution. It is RECOMMENDED to calculate rai:pop as the average within the Sample footprint. For details, see 10.48690/1531770. |
| rai:admin0 | String | yes | The administrative boundary at country level, where the Sample is located. For details, see 10.1371/journal.pone.0231866. |
| rai:admin1 | String | yes | The administrative boundary at districts level, where the Sample is located. For details, see 10.1371/journal.pone.0231866. |
| rai:admin2 | String | yes | The administrative boundary at municipality level, where the Sample is located. For details, see 10.1371/journal.pone.0231866. |
The taco-toolbox automatically generates the rai fields based on the centroid field. It is required to count with a Google Earth Engine account to access the data.
The Data Pile (DATA)
The items are serialized as a single BLOB. Each item is organized based on the FOOTER.
File Extension
The file extension MUST be .tortilla for a single Tortilla file. If the file is split into multiple partitions, the naming convention MUST follow the pattern ####.part.tortilla, where #### is a four-digit number representing the partition index.
Media Type
The Tortilla format uses the following proposed media type to identify files compliant with this specification: application/vnd.tortilla+blob
FAQ
Why not use Parquet?
Parquet is a column-oriented data file format optimized for efficient storage and retrieval of structured data. While some platforms, such as HuggingFace, promote storing image data within Parquet files, they mainly think around about small, RGB uint8 images. This is quite different from the detailed, high-res images often used in Earth Observation (EO).
In EO, a frequent requirement is to read small portions of an image (commonly referred to as partial reads), and this typically happens without users even noticing (e.g., check any raster plotting function in your favorite programming language). Spatial partial reads are straightforward with formats like GeoTIFF, where the byte location of spatial chunks is stored in the file header. This enables direct access to specific parts of the image without loading the entire file.
However, when multiple images are serialized into a column chunk of a Parquet file, the header location for these images becomes inaccessible to the data user. As a result, users must load into memory the entire chunk to access any part of the images, which is highly inefficient for large EO images. Without support for partial reads, the images lose their cloud-optimized properties, even if the original format was COG.
In constrast, Tortilla internally use Parquet ONLY for its primary purpose: managing number and string data. By embedding all metadata in the Parquet FOOTER, Tortilla allows efficient, streamlined access to the entire metadata with a single request. Image data, on the other hand, is stored separately in a Binary Large Object (BLOB) format within the DATA pile (see the Format section). Leveraging GDAL’s Virtual File System (VFS), Tortilla provides an advanced indexing system that ensures all samples retain their cloud-optimized functionality. This design enables seamless, scalable workflows that go beyond what Parquet alone can support.
Why not use Zarr?
We encourage data providers to consider using Zarr as it flexibility can be useful for some use cases. There are three key differences between Zarr and Tortilla:
-
Tortilla is a
Collectionformat while Zarr is an DataCube format. A Tortilla file is simple a collection of objects which share a common metadata structure. A sample can be an image, vector, or any object that can be serialized into a binary format. The data and metadata are kept entirely separate. The metadata is stored in a Parquet file, while the data is stored in a BLOB. The Tortilla format enables users to access all metadata by a ONE request, regardless of the dataset size. This is not possible with Zarr, where metadata is scattered throughout its tree structure in JSON files. -
Tortilla is a portable and self-contained file format, whereas Zarr is not. A Tortilla file is a single file. Although it can be split into multiple partitions, each partition is independently readable. In contrast, A Zarr object is organized in a FOLDER tree structure, with
metadatastored in JSON files anddatain binary files. The data is chunked into smaller pieces (defined by the data provider), which are stored in separate files. As a result, a Zarr object can potentially contain hundreds, thousands, or millions of files, depending on the chunking strategy and data volume.
All the Tortillas share some common characteristics, while Zarr objects does not provide a standard way to store metadata. The Tortilla format is created specifically for ready-to-use Earth Observation machine learning datasets, as we do not try to be a general-purpose format, it permits us to force data providers to follow a standard, which is essential for the interoperability of datasets.
- Tortilla is better suited for static datasets, while Zarr is more appropriate for dynamic datasets. The structure of Tortilla makes appending or modifying data after the file is created a challenging task. While editing is possible using the taco-toolbox, it can be slow and may require significant memory and storage, especially with large datasets. In contrast, Zarr is designed to efficiently handle continuous growth and shrinkage, making it a better choice for dynamic datasets.
Can I save 4-D or 5-D tensor in Tortilla?
Yes, but probably GeoTIFF would not be an ideal choice. You can try NetCDF or HDF5. You can also explore nested Tortilla structures. See the taco-toolbox for more information.
Why am I not getting expected performance in online mode with the tortilla readers?
You’re likely making several independent byte-range requests.
# Independent requests are slower due to multiple network calls
for offset, length in ranges:
headers = {"Range": f"bytes={offset}-{offset+length-1}"}
response = requests.get(url, headers=headers)
# Process each response
# ...
The ideal approach is to combine all byte ranges into a single request.
# Combine byte ranges into a Range header format
range_header = ",".join([f"bytes={offset}-{offset+length-1}" for offset, length in ranges])
# Send a single request with multiple byte ranges
headers = {"Range": range_header}
response = requests.get(url, headers=headers)
While we can provide better ways to abstract this for users, we believe that the online mode should be used only to get glimpses of the data and not for complex operations that could overload the server. For machine learning or other complex tasks, the ideal approach is to download the entire dataset or a specific portion of it (see examples) and perform operations locally.
What Does the Tortilla Reader Do?
The Tortilla reader, whether in R, Julia, or Python, first loads the FOOTER bytes (i.e. parquet file) into memory. It then converts the FOOTER into a DataFrame object. This DataFrame includes the internal:subfile column, which stores snippets of GDAL Virtual File Systems for efficient access to each item. The /vsisubfile/ handler enables access to specific binary chunks, while /vsicurl/ is used to handle online files.